So I have a conversion file for Google Drive docx to Logseq .md files.
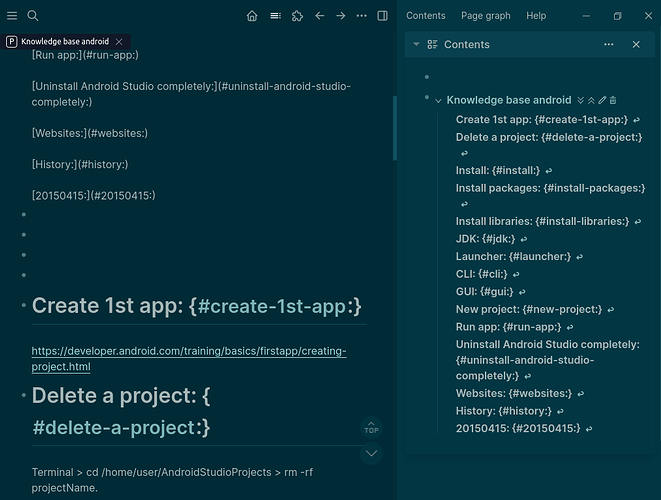
Logseq desktop 0.10.9 with plugin logseq-toc-plugin v2.0.3 outputs a left column with TOC with headers separated with individual links, correctly linking to the main page’s headers. Incorrectly the TOC headers also include the text below the headers.
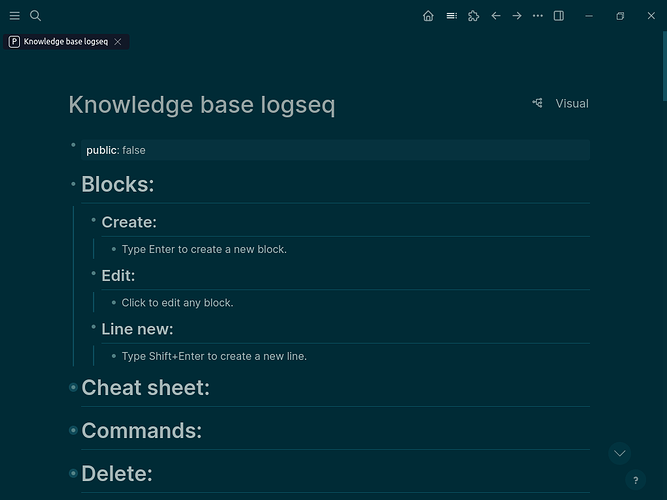
The main page correctly shows no TOC and does show headers with text below headers.
I once had the left column TOC correctly showing only headers with separated and individual links to the main page headers, but cannot replicate this perfect scenario.
Here’s a bug report I generated with AI:
Logseq Bug Report: Left Sidebar TOC Displays Incorrect (Stale/Original) Header Text
Problem Description: When importing a Markdown file, the Logseq left sidebar Table of Contents (TOC) incorrectly displays header text with remnants of original page numbers (e.g., “Header: 5”) even though the source Markdown file contains clean, standard Markdown headers (e.g., “# Header”). The main page content itself is rendered correctly, and TOC links are functional.
Environment:
- Operating System: Raspberry Pi OS (Debian based)
- Logseq Version: Logseq Flatpak version 0.10.9
- Sync Method: Nextcloud (graph folder synced via Nextcloud client)
- Conversion Tools:
- Pandoc (Version 3.1.11.1, as seen in
pandoc --version output)
- Custom Python script (
clean_and_format_md.py)
Steps to Reproduce:
- Source Document: Start with a Microsoft Word (
.docx) document that contains a table of contents generated by Word, where headers are followed by page numbers (e.g., “Section Title … 5”).
- (Example snippet from initial Pandoc output illustrating the problem source): `## $
$
backup: 5$
$
CLI: 5$`
- Conversion to Raw Markdown (Pandoc): Use Pandoc to convert the
.docx file to raw Markdown:Bashpandoc "input.docx" -f docx -t markdown --wrap=none --markdown-headings=atx -o "/tmp/raw.md"
- Resulting raw Markdown will contain lines like: `Markdown##
backup: 5
CLI: 5
GUI: 6
backup
Best is GUI.
CLI`(Note: Pandoc duplicates headings in a TOC-like structure and also creates actual content headings.)
- Clean and Format Markdown (Custom Python Script): Use a custom Python script to clean the raw Markdown:
- Removes all Pandoc-generated internal TOC links (e.g.,
[**backup: 5**](#backup) lines).
- Cleans actual content headers by removing any trailing
: page_number suffixes, bolding (**), and blockquote markers (>).
- Ensures consistent blank line spacing.
- Outputs the cleaned Markdown to a file with a hyphenated, all-lowercase filename (e.g.,
knowledge-base-raspberry-pi.md).
- (Crucially, the
alias:: property was removed from the Python script to resolve prior “two files” issues in Logseq.)**Relevant Python script logic snippet for header cleaning:Python# ... inside the cleaning loop ... if header_match: hashes = header_match.group(1) header_text = header_match.group(2).strip() header_text = re.sub(r':\s*\d*\s*Example of the final cleaned Markdown output (as confirmed by cat -A):`Markdown# backup$
$
Best is GUI.$
$
CLI$
$
Raspberry Pi → remove SD card …$
$
GUI$
$
RaPi5 → SD Card Copier …$
$
bluetooth$
$
Raspberry Pi → insert cabled keyboard …$*(Note the absence of : $ after headers and the general cleanliness of the Markdown.)* 4. **Aggressive Logseq Cache/Index Cleanup:** Before placing the file, ensure Logseq's environment is clean:Bashflatpak kill com.logseq.Logseq # Or kill your Logseq process
sleep 3
rm -rf ~/.config/Logseq
rm -rf ~/.cache/Logseq
rm -rf ~/.var/app/com.logseq.Logseq/config/Logseq/
rm -rf ~/.var/app/com.logseq.Logseq/cache/
rm -rf “/path/to/your/logseq-private/.logseq/” # Delete graph’s internal index5. **Place Cleaned Markdown File:** Ensure no old versions of the file exist in the Logseqpages/directory. Then, place the **single, cleaned Markdown file** (e.g.,/home/rapi5/Nextcloud/logseq-private/pages/knowledge-base-raspberry-pi.md) into the pages/` folder of your Logseq graph.
6. Launch Logseq: Start Logseq. It will perform a full re-index.
Observed Behavior:
- File Management: Logseq correctly identifies and loads only one file named
knowledge-base-raspberry-pi.
- Main Page Content: The main content area of the
knowledge-base-raspberry-pi page renders perfectly. It displays the cleaned headers and text, with no embedded Table of Contents at the top.
- Left Sidebar TOC:
- The TOC correctly identifies and separates individual headers (e.g., “backup”, “CLI”, “GUI”).
- The links within the TOC are functional and correctly navigate to the corresponding sections on the main page.
- However, the display text for each TOC entry in the left sidebar still includes the original page numbers and formatting (e.g., “backup: 5”, “CLI: 5”, “Browser 6”, “Firefox 7”, “Bookmarks 7”, “Import 7”). This is despite the underlying Markdown file containing only the clean header text (e.g.,
# backup, ## CLI).
Expected Behavior:
The left sidebar Table of Contents should accurately reflect the cleaned header text present in the Markdown file. For example, # backup should display as “backup” in the TOC, not “backup: 5”.
Impact: While the main page is clean and navigation works, the cluttered TOC sidebar can be confusing and less useful for quick scanning of topics. It suggests that Logseq might retain or infer metadata from the original document source (via Pandoc’s initial output) for TOC rendering, even when the Markdown file itself has been thoroughly cleaned., ‘’, header_text) # Removes “: 5”
header_text = header_text.replace(‘*’, ‘’) # Removes bolding
current_block = [f"{hashes} {header_text}"]
…*Example of the **final cleaned Markdown output** (as confirmed by DISCOURSE_PLACEHOLDER_16):*DISCOURSE_PLACEHOLDER_17*(Note the absence of DISCOURSE_PLACEHOLDER_18` after headers and the general cleanliness of the Markdown.)*
- Aggressive Logseq Cache/Index Cleanup: Before placing the file, ensure Logseq’s environment is clean:
DISCOURSE_PLACEHOLDER_19
- Place Cleaned Markdown File: Ensure no old versions of the file exist in the Logseq
DISCOURSE_PLACEHOLDER_20 directory. Then, place the single, cleaned Markdown file (e.g., DISCOURSE_PLACEHOLDER_21) into the DISCOURSE_PLACEHOLDER_22 folder of your Logseq graph.
- Launch Logseq: Start Logseq. It will perform a full re-index.
Observed Behavior:
- File Management: Logseq correctly identifies and loads only one file named
DISCOURSE_PLACEHOLDER_23.
- Main Page Content: The main content area of the
DISCOURSE_PLACEHOLDER_24 page renders perfectly. It displays the cleaned headers and text, with no embedded Table of Contents at the top.
- Left Sidebar TOC:
- The TOC correctly identifies and separates individual headers (e.g., “backup”, “CLI”, “GUI”).
- The links within the TOC are functional and correctly navigate to the corresponding sections on the main page.
- However, the display text for each TOC entry in the left sidebar still includes the original page numbers and formatting (e.g., “backup: 5”, “CLI: 5”, “Browser 6”, “Firefox 7”, “Bookmarks 7”, “Import 7”). This is despite the underlying Markdown file containing only the clean header text (e.g.,
DISCOURSE_PLACEHOLDER_25, DISCOURSE_PLACEHOLDER_26).
Expected Behavior:
The left sidebar Table of Contents should accurately reflect the cleaned header text present in the Markdown file. For example, DISCOURSE_PLACEHOLDER_27 should display as “backup” in the TOC, not “backup: 5”.
Impact: While the main page is clean and navigation works, the cluttered TOC sidebar can be confusing and less useful for quick scanning of topics. It suggests that Logseq might retain or infer metadata from the original document source (via Pandoc’s initial output) for TOC rendering, even when the Markdown file itself has been thoroughly cleaned.
Here’s the code I run:
rapi5 raspberrypi ~ Desktop $ cat clean_and_format_md.py
# clean_and_format_md.py - Version 12.3: Remove Alias Property
import sys
import re
def clean_and_format_markdown(input_file_path, output_file_path, base_name):
"""
Reads raw Pandoc Markdown.
- Filters out all Pandoc-generated internal TOC links for a clean main page.
- Cleans up actual content headers to be concise for Logseq's native TOC.
- (Temporarily) Removes the alias property to debug "two files" issue.
- Refined blank line management for consistent header hierarchy parsing.
"""
internal_link_pattern = re.compile(r'\[.*?\]\(#.*?\)')
empty_header_pattern = re.compile(r'^\s*#+\s*$', re.IGNORECASE)
empty_blockquote_pattern = re.compile(r'^\s*>\s*$', re.IGNORECASE)
markdown_header_capture_pattern = re.compile(r'^(#+)\s*(.*)$')
try:
with open(input_file_path, 'r', encoding='utf-8') as f:
raw_lines = f.readlines()
final_output_blocks = []
current_block = []
# --- ALIAS BLOCK IS REMOVED IN THIS VERSION ---
for i, line in enumerate(raw_lines):
stripped_line = line.strip()
# Filter out unwanted lines
if internal_link_pattern.search(line) or \
(i == 0 and empty_header_pattern.fullmatch(stripped_line)) or \
empty_blockquote_pattern.fullmatch(stripped_line):
continue
if stripped_line: # Line has content
header_match = markdown_header_capture_pattern.match(stripped_line)
if header_match: # This is a Markdown header
if current_block:
final_output_blocks.append(current_block)
hashes = header_match.group(1)
header_text = header_match.group(2).strip()
header_text = re.sub(r':\s*\d*\s*$', '', header_text)
header_text = header_text.replace('*', '')
current_block = [f"{hashes} {header_text}"]
else: # Regular content line
current_block.append(stripped_line)
else: # Empty line
if current_block:
final_output_blocks.append(current_block)
current_block = []
if current_block:
final_output_blocks.append(current_block)
output_lines_flat = []
for j, block in enumerate(final_output_blocks):
if j > 0:
output_lines_flat.append("")
for line_in_block in block:
output_lines_flat.append(line_in_block)
final_output_text = "\n".join(output_lines_flat).strip() + "\n"
if final_output_text == "\n":
final_output_text = ""
with open(output_file_path, 'w', encoding='utf-8') as f:
f.write(final_output_text)
except Exception as e:
print(f"Error during Markdown cleaning and formatting: {e}", file=sys.stderr)
sys.exit(1)
if __name__ == "__main__":
if len(sys.argv) < 4:
print("Usage: python3 clean_and_format_md.py <input_raw_md_file> <output_final_md_file> <base_name>", file=sys.stderr)
sys.exit(1)
input_file = sys.argv[1]
output_file = sys.argv[2]
doc_base_name = sys.argv[3]
clean_and_format_markdown(input_file, output_file, doc_base_name)
rapi5 raspberrypi ~ Desktop $ cat docx_to_md_logseq.sh
#!/bin/bash
# Define input and output paths
INPUT_FILE="/home/rapi5/Downloads/google_drive_backup/Ken's folder 😊/House 🏠/Office/Network/Device 13 PC Raspberry Pi/Knowledge base Raspberry Pi.docx"
# --- CRITICAL CHANGE: New, unambiguous output filename ---
# Use all lowercase, hyphens instead of spaces. This is highly recommended for Logseq page names.
FINAL_OUTPUT_FILE="/home/rapi5/Nextcloud/logseq-private/pages/knowledge-base-raspberry-pi.md"
LOCAL_TEMP_DIR="/tmp" # Use a local temporary directory for processing
LOCAL_RAW_PANDOC_MD="${LOCAL_TEMP_DIR}/knowledge_base_raspberry_pi_raw.md" # Raw Pandoc output (temp file, doesn't need to change)
PYTHON_SCRIPT_PATH="/home/rapi5/Desktop/clean_and_format_md.py" # Path to the new Python script (Version 12.2)
# Ensure pandoc, python3, rsync, dos2unix are installed (removed checks for brevity here, assume they are in your actual script)
# Clean up any previous temp files before starting
rm -f "$LOCAL_RAW_PANDOC_MD"
# Get base name for Logseq page title and aliases
# This will be "Knowledge base Raspberry Pi"
BASE_NAME=$(basename "$INPUT_FILE" .docx)
# --- STEP 1: Convert DOCX to Markdown to local temp file: $LOCAL_RAW_PANDOC_MD ---
echo "--- STEP 1: Converting DOCX to Markdown to local temp file: $LOCAL_RAW_PANDOC_MD ---"
pandoc "$INPUT_FILE" \
-f docx \
-t markdown \
--wrap=none \
--markdown-headings=atx \
-o "$LOCAL_RAW_PANDOC_MD"
if [ $? -eq 0 ]; then
echo "Conversion successful: '$INPUT_FILE' converted to '$LOCAL_RAW_PANDOC_MD'"
else
echo "Error during pandoc conversion."
exit 1
fi
echo "--- STEP 1 Complete ---"
# --- INTERMEDIATE CHECK 1: Raw Pandoc output (first 50 lines) ---
echo "--- INTERMEDIATE CHECK 1: Raw Pandoc output in $LOCAL_RAW_PANDOC_MD (showing all characters) ---"
cat -A "$LOCAL_RAW_PANDOC_MD" | head -n 50
echo "--- END INTERMEDIATE CHECK 1 ---"
# --- STEP 2: Clean and format Markdown using Python script ---
echo "--- STEP 2: Cleaning and formatting Markdown using Python script ---"
# Convert to Unix line endings *before* Python processes, just in case
dos2unix "$LOCAL_RAW_PANDOC_MD"
# Pass the base name to the Python script. The Python script will handle cleaning it for the alias.
python3 "$PYTHON_SCRIPT_PATH" "$LOCAL_RAW_PANDOC_MD" "$FINAL_OUTPUT_FILE" "$BASE_NAME"
if [ $? -ne 0 ]; then
echo "Error: Python cleaning and formatting script failed."
exit 1
fi
echo "--- STEP 2 Complete ---"
# --- INTERMEDIATE CHECK 2: Final output file after Python (first 50 lines) ---
echo "--- INTERMEDIATE CHECK 2: Final output file $FINAL_OUTPUT_FILE (showing all characters) ---"
cat -A "$FINAL_OUTPUT_FILE" | head -n 50
echo "--- END INTERMEDIATE CHECK 2 ---"
# Give Nextcloud a moment to catch up and force a disk sync
echo "--- Pausing for 10 seconds to allow Nextcloud to sync and forcing disk sync... ---"
sleep 10
sync # Force writes to disk
echo "--- Pause complete. ---"
# Clean up local temporary files
rm -f "$LOCAL_RAW_PANDOC_MD"
echo "All conversion and cleanup steps completed. Final File: $FINAL_OUTPUT_FILE"
# Logseq specific considerations:
echo "Remember to re-index your Logseq graph or restart Logseq to ensure the new file is loaded correctly."
echo "If issues persist, clear Logseq's cache as described in the instructions."